Update 6/23/23: Liu and coauthors have just formally described the order Xenasmatellales, based on analyses of nSSU, ITS, nLSU, mtSSU, tef1α, rpb1 and rpb2 sequences. When the original post below was written, back in 2017, there was only one Xenasmatella genome on MycoCosm, X. vaga. Today, there is also a genome of X. tulasnelloidea. The sister group of the Xenasma clade in the MycoCosm genome tree of Agricomycetes is “Phlebia subcretacea.” However, Phlebia is a genus of the Meruliaceae, Polyporales. Assuming that the genome strain was accurately identified (which is quite likely, since Otto Miettinen is a PI for this genome), another name will be needed. According to MycoBank, the oldest name for this species is Jacksonomyces subcretaceus (Litsch.) Jülich, Persoonia 11 (4): 427 (1982) [MB#110743]. Perhaps the next classification of Xenasmatellales will include Jacksonomyces?
This fine study highlights the glacial pace of taxonomic revision, and the disconnect between advances in phylogenetic knowledge and updates in formal classifications. Liu et al. have done beautiful, careful work on this “new” order of Agaricomycetes, with extensive sampling of Xenasmatella. Their analysis of nSSU, ITS, nLSU and mtSSU sequences includes over 40 individuals, representing approximately twelve species (see their fig. 3). Still, it is too bad that there was a seven-year lag between the discovery that Xenasmatella represents a unique clade that cannot be placed in any other order of Agaricomycetes (based on the MycoCosm phylogeny) and its formal recognition as a taxon. Automated approaches to translating phylogenies into taxonomy are still needed to communicate the latest advances in reconstructing the fungal tree of life in the form of classifications.
Original post (7/13/2017): The genome-based phylogeny on the Joint Genome Institute’s MycoCosm site suggests that Xenasmatella vaga represents a lineage that is not nested within any other recognized “order” of Agaricomycetes. JGI now hosts 188 species of Agaricomycetes, representing eighteen orders, so sampling is not a major concern. In the JGI tree, Xenasmatella vaga is placed as the sister group of a large clade containing Polyporales, Agaricomycetidae, Russulales, Thelephorales, and other clades. If this is correct, the Xenasmatella vaga genome could provide important data that would address the ecological capabilities of deep backbone nodes in Agaricomycetes. This genome was produced under the auspices of the 1000 Fungal Genomes Project, with Joey Spatafora and Jon Magnuson as the PIs for the species.
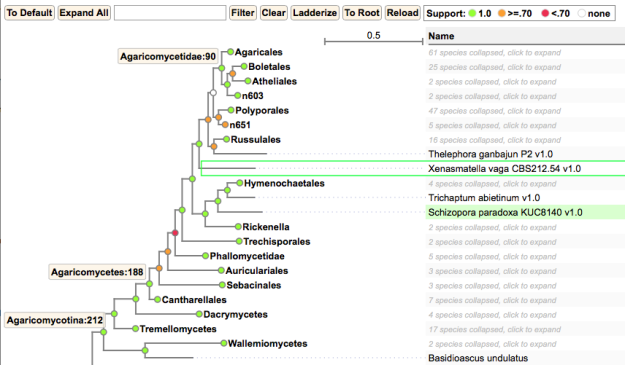
Portion of the JGI fungal phylogeny showing placement of X. vaga among the Agaricomycetes. Note support values.
Xenasmatella vaga is a representative of the “corticioid” fungi, an understudied group of Agaricomycetes with simple, resupinate fruiting bodies, which includes saprotrophs, pathogens, and ectomycorrhizal species. There are few experts on this diverse and taxonomically challenging group, which may harbor additional undiscovered major clades of Agaricomycetes.
Species page for the X. vaga genome on MycoCosm.